Introduction
In this example, we will show how to explore the cell-cycle dataset of CCTD-U using AggMap.
The CCTD-U[1] dataset is a cell-cycle transcriptome data of U2OS cells, consisting of the expression levels of 5162 genes at 5 different cell cycle stages (G1, G1 / S, S, G2, M). This dataset was transformed using z-score standard scaling.
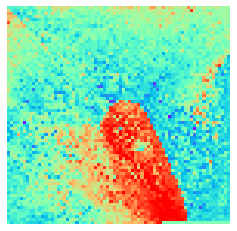
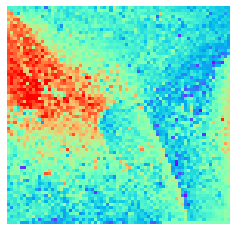
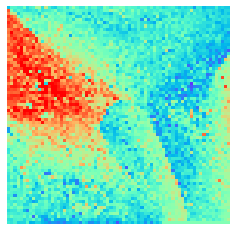
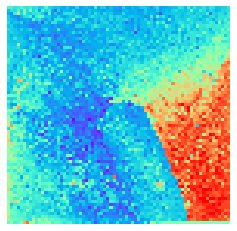
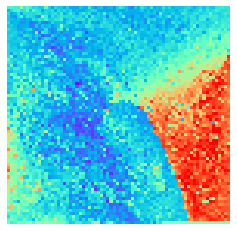
By fitting on the CCTD-U dataset, AggMap can transform it into feature maps (Fmaps). Visualization of AggMap multi-channel Fmaps of the cell-cycle CCTD-U dataset at different cell replication phases indicated that multi-channel Fmaps can easily select stage-specific genes. The stage -specific genes in the five cell-cycle phases can be easily aggregated into hot-zones in the single-channel Fmaps based on their correlations, while the multi-channel Fmaps further separate the phase-specific genes into different channels. Therefore, the multi-channel Fmaps facilitate group-specific feature learning or feature selective learning by AggMapNet.
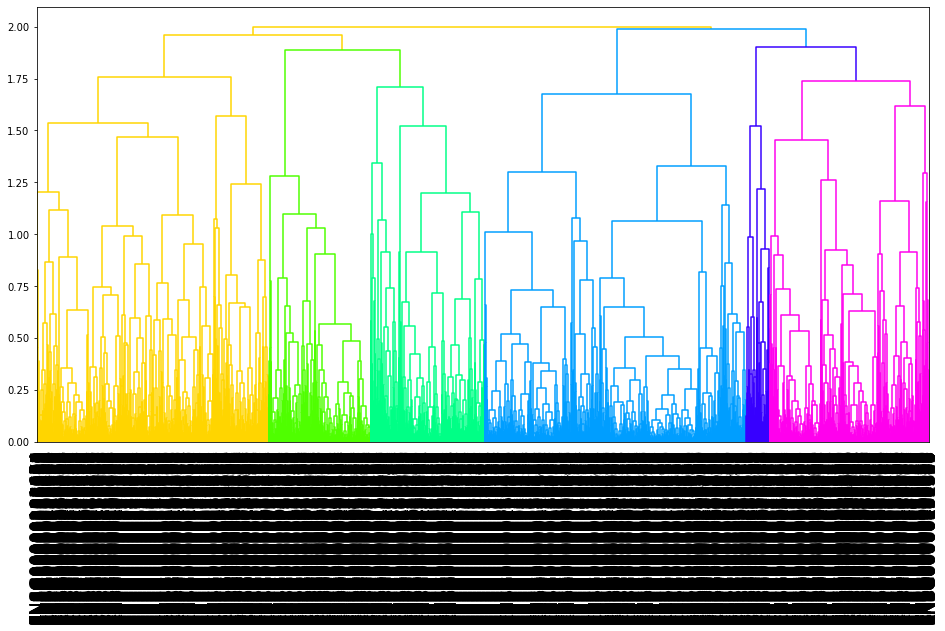
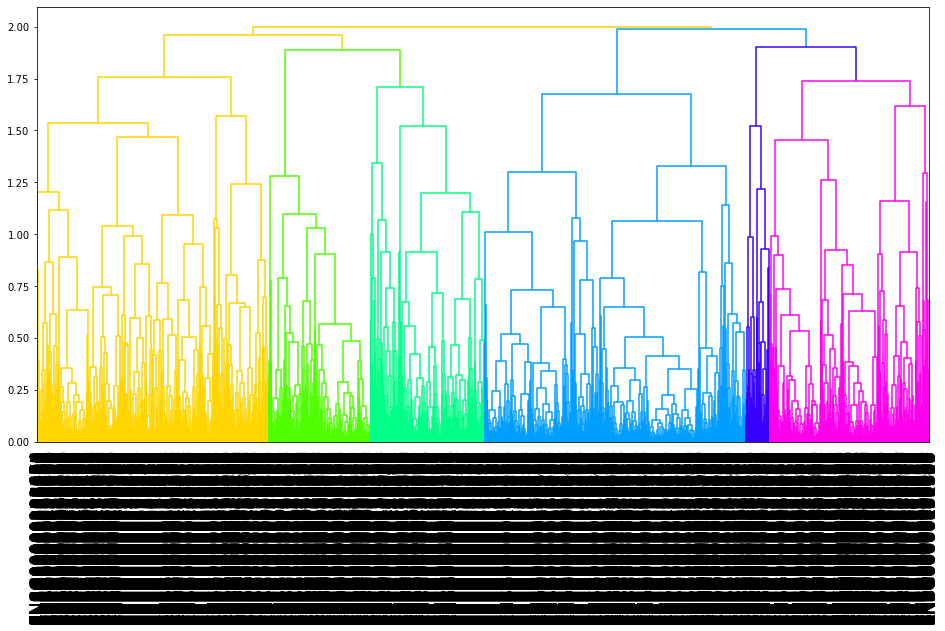
By the hierarchical clustering-guided channel splits, each cluster (group) of feature points may be separately embedded into a different Fmap channel. More clusters enable more fine-grained separations of feature points into groups.
[1]. Hao, Q., Zong, X., Sun, Q., Lin, Y.-C., Song, Y.J., Hashemikhabir, S., Hsu, R.Y., Kamran, M., Chaudhary, R. and Tripathi, V. (2020) The S-phase-induced lncRNA SUNO1 promotes cell proliferation by controlling YAP1/Hippo signaling pathway. Elife, 9, e55102.
Read data and pre-fit on AggMap
[1]:
import os
import pandas as pd
import numpy as np
import seaborn as sns
import tensorflow as tf
from sklearn.utils import shuffle
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
import matplotlib.pyplot as plt
from aggmap import AggMap, show
2022-08-01 16:25:56.434248: I tensorflow/core/util/util.cc:169] oneDNN custom operations are on. You may see slightly different numerical results due to floating-point round-off errors from different computation orders. To turn them off, set the environment variable `TF_ENABLE_ONEDNN_OPTS=0`.
[2]:
df = pd.read_excel('https://cdn.elifesciences.org/articles/55102/elife-55102-supp5-v1.xlsx')
[3]:
df = df.set_index('ensemble_id')
[4]:
dfx = df[df.columns[4:]].T
[5]:
mp = AggMap(dfx, metric='correlation', by_scipy=True)
2022-08-01 16:25:58,556 - INFO - [bidd-aggmap] - Calculating distance ...
100%|###########################################################################################################################################| 5162/5162 [00:00<00:00, 5831.88it/s]
[6]:
mp = mp.fit(cluster_channels=6, n_neighbors = 500, spread=2) #
2022-08-01 16:25:59,705 - INFO - [bidd-aggmap] - applying hierarchical clustering to obtain group information ...
UMAP(a=None, angular_rp_forest=False, b=None, init='spectral',
learning_rate=1.0, local_connectivity=1.0, metric='precomputed',
metric_kwds=None, min_dist=0.1, n_components=2, n_epochs=None,
n_neighbors=500, negative_sample_rate=5, random_state=32,
repulsion_strength=1.0, set_op_mix_ratio=1.0, spread=2,
target_metric='categorical', target_metric_kwds=None,
target_n_neighbors=-1, target_weight=0.5, transform_queue_size=4.0,
transform_seed=42, verbose=2)
Construct fuzzy simplicial set
Mon Aug 1 16:26:00 2022 Finding Nearest Neighbors
Mon Aug 1 16:26:01 2022 Finished Nearest Neighbor Search
Mon Aug 1 16:26:03 2022 Construct embedding
completed 0 / 500 epochs
completed 50 / 500 epochs
completed 100 / 500 epochs
completed 150 / 500 epochs
completed 200 / 500 epochs
completed 250 / 500 epochs
completed 300 / 500 epochs
completed 350 / 500 epochs
completed 400 / 500 epochs
completed 450 / 500 epochs
Mon Aug 1 16:26:11 2022 Finished embedding
2022-08-01 16:26:11,784 - INFO - [bidd-aggmap] - Applying grid assignment of feature points, this may take several minutes(1~30 min)
2022-08-01 16:26:56,509 - INFO - [bidd-aggmap] - Finished
Transform the data by AggMap
[7]:
X = mp.batch_transform(dfx.values, scale_method='standard')
100%|###################################################################################################################################################| 9/9 [00:01<00:00, 5.74it/s]
Multi-channel Fmaps
[8]:
for i in range(9):
show.imshow_wrap(X[i], color_list=mp.colormaps.values(), x_max=1, vmin=-0.5, vmax=2)
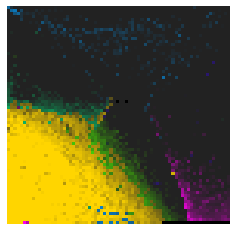


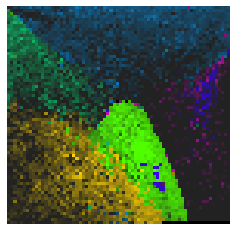



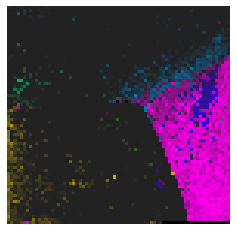
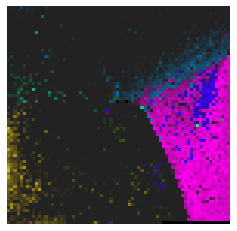
Single-channel Fmaps
[9]:
for i in range(9):
fig, ax = plt.subplots(figsize=(4,4))
sns.heatmap(X[i].sum(axis=-1).reshape(*mp.fmap_shape), ax = ax, cmap = 'rainbow', yticklabels=False, xticklabels=False, cbar=False, vmin=-2, vmax=2)
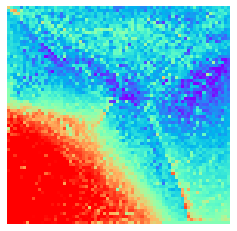
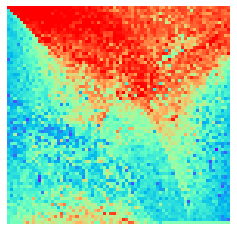
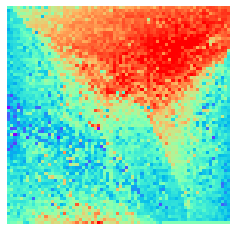
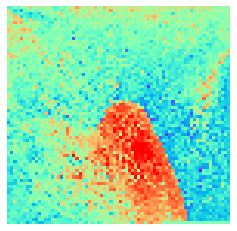
[10]:
fig, ax = plt.subplots(figsize=(8,10))
sns.heatmap(X[i].sum(axis=-1).reshape(*mp.fmap_shape), ax = ax, cmap = 'rainbow',
yticklabels=False, xticklabels=False, cbar=True, vmin=-2, vmax=2,
cbar_kws = dict(use_gridspec=False,location="top"))
fig.savefig('legend.pdf')
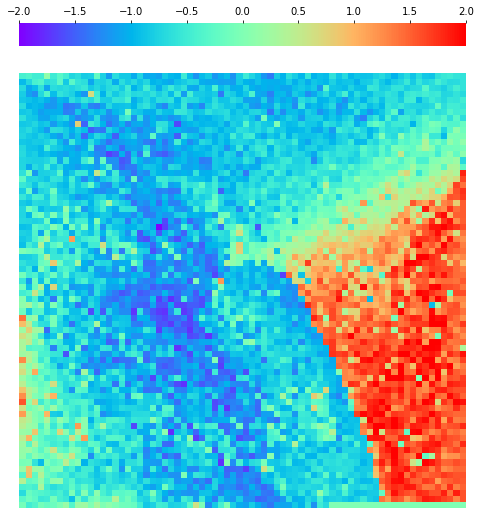
Visulization
01.scatter plot
[11]:
mp.plot_scatter()
2022-08-01 16:26:59,451 - INFO - [bidd-aggmap] - generate file: ./feature points_5162_correlation_umap_scatter
2022-08-01 16:26:59,480 - INFO - [bidd-aggmap] - save html file to ./feature points_5162_correlation_umap_scatter
[11]:
02.grid plot
[12]:
mp.plot_grid()
2022-08-01 16:26:59,508 - INFO - [bidd-aggmap] - generate file: ./feature points_5162_correlation_umap_mp
2022-08-01 16:26:59,530 - INFO - [bidd-aggmap] - save html file to ./feature points_5162_correlation_umap_mp
[12]:
03.tree plot
[15]:
mp.to_nwk_tree()
[15]:
| TYPE | colors | STYLE | |
|---|---|---|---|
| ENSG00000003436 | clade | #ffd500 | normal |
| ENSG00000004139 | clade | #ffd500 | normal |
| ENSG00000004799 | clade | #ffd500 | normal |
| ENSG00000004948 | clade | #ffd500 | normal |
| ENSG00000005073 | clade | #ffd500 | normal |
| ... | ... | ... | ... |
| ENSG00000280053 | clade | #ff00ed | normal |
| ENSG00000280202 | clade | #ff00ed | normal |
| ENSG00000280206 | clade | #ff00ed | normal |
| ENSG00000283154 | clade | #ff00ed | normal |
| ENSG00000285530 | clade | #ff00ed | normal |
5162 rows × 3 columns
[14]:
mp.plot_tree()
[14]:
[ ]: