3. Training the CRC detection models based on MEGMA Fmaps
Now that we have fitted the megma object for the Fmaps generation, we can train the CRC detection models based on these transformed Fmaps. We are going to use the convolutional neural network (CNN) based AggMapNet to train the Fmaps. AggMapNet is a simple yet efficient network architecture for low-sample size data, it has fewer trainable parameters and early-stopping strategy to stop from overfitting.
3.1 Training & test AggMapNet on overall MEGMA Fmaps
3.2 Training and test AggMapNet on country specific MEGMA Fmaps
3.3 Comparing the STST performance and discussion
[8]:
from sklearn.metrics import confusion_matrix
from sklearn.metrics import precision_recall_curve
from sklearn.metrics import roc_auc_score
from sklearn.metrics import auc as calculate_auc
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import numpy as np
import os
from aggmap import AggMap, AggMapNet
from aggmap import show, loadmap
np.random.seed(666) #just for reaptable results
[9]:
## AggMapNet parameters
epochs = 30 #number of epochs
lr = 1e-4 #learning rate
batch_size = 1 #batch size
conv1_kernel_size = 7 #kernal size of the first cnn layer
gpuid = 1 #use which gpu to train the model
def train(megma, train_country, model_save_dir):
## data process parameters
minv = 1e-8 #minimal value for log-transform
scale_method = 'standard' #data scaling method
url = 'https://raw.githubusercontent.com/shenwanxiang/bidd-aggmap/master/docs/source/_example_MEGMA/dataset/'
dfx_vector = pd.read_csv(url + '%s_dfx.csv' % train_country, index_col='Sample_ID')
dfx_vector = np.log(dfx_vector + minv)
dfy = pd.read_csv(url + '%s_dfy.csv' % train_country, index_col='Sample_ID')
X = megma.batch_transform(dfx_vector.values,
scale_method = scale_method)
Y = pd.get_dummies(dfy.Group).values
# fit AggMapNet
clf = AggMapNet.MultiClassEstimator(epochs = epochs,
batch_size = batch_size,
lr = lr,
conv1_kernel_size = conv1_kernel_size,
gpuid = gpuid,
verbose = 0)
clf.fit(X, Y)
## save supervised AggMapNet model
model_save_name = os.path.join(model_save_dir, 'aggmapnet.%s' % train_country)
clf.save_model(model_save_name)
return clf
def test(clf, megma, test_country):
## data process parameters
minv = 1e-8 #minimal value for log-transform
scale_method = 'standard' #data scaling method
url = 'https://raw.githubusercontent.com/shenwanxiang/bidd-aggmap/master/docs/source/_example_MEGMA/dataset/'
dfx_vector = pd.read_csv(url + '%s_dfx.csv' % test_country, index_col='Sample_ID')
dfx_vector = np.log(dfx_vector + minv)
dfy = pd.read_csv(url + '%s_dfy.csv' % test_country, index_col='Sample_ID')
testX = megma.batch_transform(dfx_vector.values,
scale_method = scale_method)
testY = pd.get_dummies(dfy.Group).values
y_true = testY[:,0]
y_pred = clf.predict(testX)[:,0]
y_score = clf.predict_proba(testX)[:,0]
tn, fp, fn, tp = confusion_matrix(y_true, y_pred).ravel()
acc = (tp + tn) / sum([tn, fp, fn, tp])
sensitivity = tp / sum([tp, fn])
specificity = tn / sum([tn, fp])
roc_auc = roc_auc_score(y_true, y_score)
precision = tp / sum([tp, fp])
recall = tp / sum([tp, fn]) #equals to sensitivity
res = {'test_country':test_country,
'accuracy':acc,
'roc_auc':roc_auc,
'sensitivity': sensitivity,
'specificity': specificity,
'precision':precision,
'recall':recall}
return res
def get_non_diag_mean(df):
v = df.values.copy()
np.fill_diagonal(v, np.nan)
mean = pd.DataFrame(v, columns = df.columns, index = df.index).mean(axis=1).round(2)
std = pd.DataFrame(v, columns = df.columns, index = df.index).std(axis=1).round(2)
return mean,std
3.1 Training and test AggMapNet on overall MEGMA Fmaps
In this section, we will introduce how to employ the CNN-based AggMapNet to train CRC detection models based on the megma_all generated 2D-microbiomeprints (Fmaps). The CRC dection model is a classfication model because we have the binary labels (CRCs or CTRs) in our data. Same as the Fmaps generation, we can use the study to stduy transfer(STST) method to test the performance of our model.
Please be Noted that in the STST experiment, the Fmaps can be transformed by a country-specific megma (fitted or trained by one country unlabelled metagenomic data, such as megma_usa) or an overall megma (fitted or trained by all unlabelled metagenomic data, i.e., the megma_all), becuase MEGMA is an unsupervised learning method and only needs to be fitted on the unlabelled data.
First of all, let’s try to explore the STST model performance on the Fmaps transformed by an overall megma_all . Specifically, we are going to train the CRC classficaition model AggMapNet (supervised model) based the Fmaps that generated by megma_all (unsupervised model). We will train the CRC detection model based on one country data and test the model performance on the rest of the countries.
[10]:
save_dir = './megma_overall_model'
if not os.path.exists(save_dir):
os.makedirs(save_dir)
# load the pre-fitted megma_all object
megma = loadmap('./megma/megma.all')
countries = ['USA', 'CHN', 'DEU', 'FRA', 'AUS', ]
all_res1 = []
for train_country in countries:
clf = train(megma, train_country, save_dir,)
# Test model performance using STST strategy
for test_country in countries:
res = test(clf, megma, test_country)
res.update({'train_country':train_country})
all_res1.append(res)
df1 = pd.DataFrame(all_res1)
dfres1 = pd.crosstab(index = df1.train_country,
columns= df1.test_country,
values= df1.roc_auc,
aggfunc = np.mean)
100%|#################################################################################| 104/104 [00:01<00:00, 57.13it/s]
MultiClassEstimator(batch_norm=False, batch_size=1, conv1_kernel_size=7,
dense_avf='relu', dense_layers=[128], dropout=0.0,
epochs=30, gpuid='1', last_avf='softmax',
loss='categorical_crossentropy', lr=0.0001, metric='ACC',
monitor='val_loss', n_inception=2,
name='AggMap MultiClass Estimator', patience=10000,
random_state=32, verbose=0)
saving model to ./megma_overall_model/aggmapnet.USA
100%|###############################################################################| 104/104 [00:00<00:00, 2087.23it/s]
100%|###############################################################################| 128/128 [00:00<00:00, 1917.29it/s]
100%|###############################################################################| 120/120 [00:00<00:00, 1564.03it/s]
100%|###############################################################################| 114/114 [00:00<00:00, 1686.02it/s]
100%|###############################################################################| 109/109 [00:00<00:00, 1531.23it/s]
100%|###############################################################################| 128/128 [00:00<00:00, 1937.72it/s]
MultiClassEstimator(batch_norm=False, batch_size=1, conv1_kernel_size=7,
dense_avf='relu', dense_layers=[128], dropout=0.0,
epochs=30, gpuid='1', last_avf='softmax',
loss='categorical_crossentropy', lr=0.0001, metric='ACC',
monitor='val_loss', n_inception=2,
name='AggMap MultiClass Estimator', patience=10000,
random_state=32, verbose=0)
saving model to ./megma_overall_model/aggmapnet.CHN
100%|################################################################################| 104/104 [00:00<00:00, 992.48it/s]
100%|###############################################################################| 128/128 [00:00<00:00, 2009.78it/s]
100%|###############################################################################| 120/120 [00:00<00:00, 1729.95it/s]
100%|###############################################################################| 114/114 [00:00<00:00, 1377.46it/s]
100%|###############################################################################| 109/109 [00:00<00:00, 2221.83it/s]
100%|###############################################################################| 120/120 [00:00<00:00, 1565.51it/s]
MultiClassEstimator(batch_norm=False, batch_size=1, conv1_kernel_size=7,
dense_avf='relu', dense_layers=[128], dropout=0.0,
epochs=30, gpuid='1', last_avf='softmax',
loss='categorical_crossentropy', lr=0.0001, metric='ACC',
monitor='val_loss', n_inception=2,
name='AggMap MultiClass Estimator', patience=10000,
random_state=32, verbose=0)
saving model to ./megma_overall_model/aggmapnet.DEU
100%|###############################################################################| 104/104 [00:00<00:00, 2081.35it/s]
100%|###############################################################################| 128/128 [00:00<00:00, 1916.38it/s]
100%|###############################################################################| 120/120 [00:00<00:00, 1849.31it/s]
100%|################################################################################| 114/114 [00:00<00:00, 871.78it/s]
100%|###############################################################################| 109/109 [00:00<00:00, 1574.34it/s]
100%|###############################################################################| 114/114 [00:00<00:00, 2403.50it/s]
MultiClassEstimator(batch_norm=False, batch_size=1, conv1_kernel_size=7,
dense_avf='relu', dense_layers=[128], dropout=0.0,
epochs=30, gpuid='1', last_avf='softmax',
loss='categorical_crossentropy', lr=0.0001, metric='ACC',
monitor='val_loss', n_inception=2,
name='AggMap MultiClass Estimator', patience=10000,
random_state=32, verbose=0)
saving model to ./megma_overall_model/aggmapnet.FRA
100%|###############################################################################| 104/104 [00:00<00:00, 2198.78it/s]
100%|###############################################################################| 128/128 [00:00<00:00, 1809.99it/s]
100%|###############################################################################| 120/120 [00:00<00:00, 2185.13it/s]
100%|###############################################################################| 114/114 [00:00<00:00, 2364.74it/s]
100%|###############################################################################| 109/109 [00:00<00:00, 1835.07it/s]
100%|###############################################################################| 109/109 [00:00<00:00, 1886.13it/s]
MultiClassEstimator(batch_norm=False, batch_size=1, conv1_kernel_size=7,
dense_avf='relu', dense_layers=[128], dropout=0.0,
epochs=30, gpuid='1', last_avf='softmax',
loss='categorical_crossentropy', lr=0.0001, metric='ACC',
monitor='val_loss', n_inception=2,
name='AggMap MultiClass Estimator', patience=10000,
random_state=32, verbose=0)
saving model to ./megma_overall_model/aggmapnet.AUS
100%|###############################################################################| 104/104 [00:00<00:00, 1984.11it/s]
100%|###############################################################################| 128/128 [00:00<00:00, 1904.11it/s]
100%|###############################################################################| 120/120 [00:00<00:00, 2112.85it/s]
100%|###############################################################################| 114/114 [00:00<00:00, 2035.77it/s]
100%|###############################################################################| 109/109 [00:00<00:00, 2175.88it/s]
3.2 Training and test AggMapNet on country specific MEGMA Fmaps
In this section, we will introduce how to employ the CNN-based AggMapNet to train CRC detection models using the country-specific megma generated 2D-microbiomeprints (Fmaps).
Specifically, we are going to use one country data to fit megma and aggmapnet, then we test the model performance on the rest of the countries. For example, if we trained the CRC detection model based on the USA data, we are going to use the megma_usa to transform the Fmaps for all of the countries, i.e., the country-specific megma will be used. Save as above model evaluation, we will train the AggMapNet-based CRC detection model on one country data (label data and abundance data)
and test the model performance on the rest of the countries.
[11]:
save_dir = './megma_country_model'
if not os.path.exists(save_dir):
os.makedirs(save_dir)
all_res2 = []
for train_country in countries:
# load the pre-fitted megma object., such as `megma_usa`.
megma = loadmap('./megma/megma.%s' % train_country)
clf = train(megma, train_country, save_dir)
# Test model performance using STST strategy
for test_country in countries:
res = test(clf, megma, test_country)
res.update({'train_country':train_country})
all_res2.append(res)
df2 = pd.DataFrame(all_res2)
dfres2 = pd.crosstab(index = df2.train_country,
columns= df2.test_country,
values= df2.roc_auc,
aggfunc = np.mean)
100%|###############################################################################| 104/104 [00:00<00:00, 2268.13it/s]
MultiClassEstimator(batch_norm=False, batch_size=1, conv1_kernel_size=7,
dense_avf='relu', dense_layers=[128], dropout=0.0,
epochs=30, gpuid='1', last_avf='softmax',
loss='categorical_crossentropy', lr=0.0001, metric='ACC',
monitor='val_loss', n_inception=2,
name='AggMap MultiClass Estimator', patience=10000,
random_state=32, verbose=0)
saving model to ./megma_country_model/aggmapnet.USA
100%|###############################################################################| 104/104 [00:00<00:00, 2268.67it/s]
100%|###############################################################################| 128/128 [00:00<00:00, 2426.38it/s]
100%|###############################################################################| 120/120 [00:00<00:00, 1969.87it/s]
100%|###############################################################################| 114/114 [00:00<00:00, 1809.61it/s]
100%|###############################################################################| 109/109 [00:00<00:00, 1880.95it/s]
100%|###############################################################################| 128/128 [00:00<00:00, 1946.63it/s]
MultiClassEstimator(batch_norm=False, batch_size=1, conv1_kernel_size=7,
dense_avf='relu', dense_layers=[128], dropout=0.0,
epochs=30, gpuid='1', last_avf='softmax',
loss='categorical_crossentropy', lr=0.0001, metric='ACC',
monitor='val_loss', n_inception=2,
name='AggMap MultiClass Estimator', patience=10000,
random_state=32, verbose=0)
saving model to ./megma_country_model/aggmapnet.CHN
100%|###############################################################################| 104/104 [00:00<00:00, 1767.68it/s]
100%|###############################################################################| 128/128 [00:00<00:00, 1284.44it/s]
100%|###############################################################################| 120/120 [00:00<00:00, 1990.42it/s]
100%|###############################################################################| 114/114 [00:00<00:00, 1538.51it/s]
100%|###############################################################################| 109/109 [00:00<00:00, 2608.40it/s]
100%|###############################################################################| 120/120 [00:00<00:00, 1070.05it/s]
MultiClassEstimator(batch_norm=False, batch_size=1, conv1_kernel_size=7,
dense_avf='relu', dense_layers=[128], dropout=0.0,
epochs=30, gpuid='1', last_avf='softmax',
loss='categorical_crossentropy', lr=0.0001, metric='ACC',
monitor='val_loss', n_inception=2,
name='AggMap MultiClass Estimator', patience=10000,
random_state=32, verbose=0)
saving model to ./megma_country_model/aggmapnet.DEU
100%|###############################################################################| 104/104 [00:00<00:00, 1062.87it/s]
100%|###############################################################################| 128/128 [00:00<00:00, 1088.08it/s]
100%|###############################################################################| 120/120 [00:00<00:00, 2722.76it/s]
100%|###############################################################################| 114/114 [00:00<00:00, 1996.81it/s]
100%|###############################################################################| 109/109 [00:00<00:00, 1926.46it/s]
100%|###############################################################################| 114/114 [00:00<00:00, 2088.69it/s]
MultiClassEstimator(batch_norm=False, batch_size=1, conv1_kernel_size=7,
dense_avf='relu', dense_layers=[128], dropout=0.0,
epochs=30, gpuid='1', last_avf='softmax',
loss='categorical_crossentropy', lr=0.0001, metric='ACC',
monitor='val_loss', n_inception=2,
name='AggMap MultiClass Estimator', patience=10000,
random_state=32, verbose=0)
saving model to ./megma_country_model/aggmapnet.FRA
100%|###############################################################################| 104/104 [00:00<00:00, 2141.66it/s]
100%|###############################################################################| 128/128 [00:00<00:00, 1097.15it/s]
100%|###############################################################################| 120/120 [00:00<00:00, 1602.63it/s]
100%|###############################################################################| 114/114 [00:00<00:00, 1641.69it/s]
100%|###############################################################################| 109/109 [00:00<00:00, 1642.69it/s]
100%|###############################################################################| 109/109 [00:00<00:00, 1987.70it/s]
MultiClassEstimator(batch_norm=False, batch_size=1, conv1_kernel_size=7,
dense_avf='relu', dense_layers=[128], dropout=0.0,
epochs=30, gpuid='1', last_avf='softmax',
loss='categorical_crossentropy', lr=0.0001, metric='ACC',
monitor='val_loss', n_inception=2,
name='AggMap MultiClass Estimator', patience=10000,
random_state=32, verbose=0)
saving model to ./megma_country_model/aggmapnet.AUS
100%|###############################################################################| 104/104 [00:00<00:00, 2034.28it/s]
100%|###############################################################################| 128/128 [00:00<00:00, 2132.14it/s]
100%|###############################################################################| 120/120 [00:00<00:00, 2206.96it/s]
100%|###############################################################################| 114/114 [00:00<00:00, 2569.98it/s]
100%|###############################################################################| 109/109 [00:00<00:00, 1904.41it/s]
3.3 Comparing the STST performance and discussion
Now, let’s compare the ROC-AUC performance of CRC detection model that trained on the Fmaps generated by overall megma and country-specific megma. We can see that the megma_all shows better performance than the country specific ones.
This means that the Fmaps generated by megma_all have a more reasonable feature arrangement and combination than country-specific megma, and using these megma_all Fmaps to train the detection model will achieve better performance. This result is consistent with our previous discussion: megma shows better performance under large sample fitting. Since the fitting of megma only requires unlabeled data, in practice we can use a large amount of unlabeled data to pre-train
megma, which is very helpful to the model performance.
If the unlabeled data used to fit the megma is still limited, we can also adjust the var_thr parameter in the fitting stage of megma to make the generated Fmaps more structured. In this example, we used var_thr = 3 to fit the country-specific megma for each country, but this value can be adjusted in each country, and we believe that under a larger value, such as 10, better roc-auc performances will be achieved.
[12]:
sns.set(style = 'white', font_scale=2)
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(18,6), sharex = True, sharey = True)
y1 = get_non_diag_mean(dfres1)[0].to_frame(name = 'Overall MEGMA')
y2 = get_non_diag_mean(dfres2)[0].to_frame(name = 'Country MEGMA')
y1_err = get_non_diag_mean(dfres1)[1].to_frame(name = 'Overall MEGMA')
y2_err = get_non_diag_mean(dfres2)[1].to_frame(name = 'Country MEGMA')
y = y1.join(y2)
y_err = y1_err.join(y2_err)
color = sns.color_palette("rainbow_r", 5) #PiYG
y.plot(kind = 'bar',ax = ax, color = color, yerr= y_err,ylim = (0.5, 0.96), error_kw=dict(ecolor='gray', lw=2, capsize=6, capthick=2))
ax.set_ylabel('ROC-AUC')
#ax.set_title('Study To Study Transfer (STST) Average Performance')
ax.set_xlabel('Study To Study Transfer Training Country')
for x_, y_ in enumerate(y['Overall MEGMA'].values):
ax.annotate('%.2f' % y_, xy = (x_-0.39, y_+0.002), fontsize =22)
for x_, y_ in enumerate(y['Country MEGMA'].values):
ax.annotate('%.2f' % y_, xy = (x_+0.14, y_+0.002), fontsize =22)
ax.set_xticklabels(labels = y.index,rotation=0, ha = 'center')
ax.set_ylim(0.5,1.05)
ax.tick_params(bottom='on', left='off', labelleft='on', labelbottom='on', pad=-.6,)
sns.despine(top=True, right=True, left=False, bottom=False)
fig.savefig('./images/STST_megma_all_vs_megma_country.pdf', bbox_inches='tight', dpi=400)
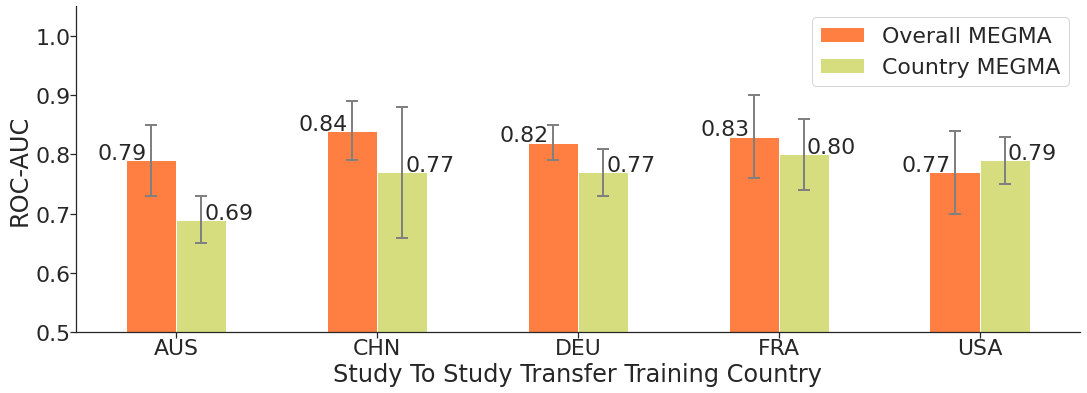
For overall megma, we can see that the model trained on the
megma_allgenerated USA Fmaps shows worst performance (STST average roc-auc: 0.77, i.e., the model performance on the rest countries).For country-specific megma, we can see that the model trained on the
megma_AUSgenerated AUS Fmaps shows worst performance (STST average roc-auc: 0.69). This means that themegmafitted by AUS abundance data is not good enough. It is likely that the abundance data of AUS is relatively special, and the sample size is too small to present the global and intrinsic correlation between microorganisms, resulting in a biased arrangement of the 2D map.
In next section, we will explain the model trained on the Fmaps generated by overall megma and country-specific megma. We can identify the important microbes that contribute to the model prediction, and generate the saliency map using the global feature importance and the microbe 2D postions in megma object.
[20]:
sns.set(style = 'white', font_scale=1.8)
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(13,4.3), sharex=True, sharey=True)
ax1, ax2 = axes
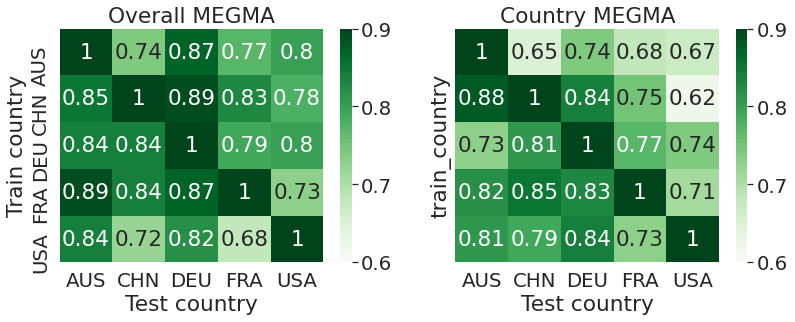
sns.heatmap(dfres1.round(2), cmap = 'Greens', ax = ax1, annot=True, vmin = 0.6, vmax = 0.9,)
ax1.set_title("Overall MEGMA")
sns.heatmap(dfres2.round(2), cmap = 'Greens', ax = ax2, annot=True, vmin = 0.6, vmax = 0.9,)
ax2.set_title("Country MEGMA")
ax1.set_ylabel('Train country')
# ax2.set_ylabel('Train country')
ax1.set_xlabel('Test country')
ax2.set_xlabel('Test country')
fig.savefig('./images/STST_roc_megma_all_vs_megma_country.png', bbox_inches='tight', dpi=400)
fig.savefig('./images/STST_roc_megma_all_vs_megma_country.pdf', bbox_inches='tight', dpi=400)
[19]:
dfres1.to_csv('./images/STST_ROC_megma_all.csv')
dfres2.to_csv('./images/STST_ROC_megma_specific.csv')