1. Introduction
1.1 MEGMA introduction
MEGMA is short for metagenomic Microbial Embedding, Grouping, and Mapping Algorithm (MEGMA) , which is a further step development of AggMap (v.1.1.7) that specific for metagenomic data learning. MEGMA is developed to transform the tabular metagenomic data into spatially-correlated color image-like 2D-representations, named as the 2D-microbiomeprints (3D tensor data in the form of row, column and channel, or feature maps, Fmaps). 2D-microbiomeprints are multichannel feature maps (Fmaps) and are the inputs of ConvNet-based AggMapNet models.
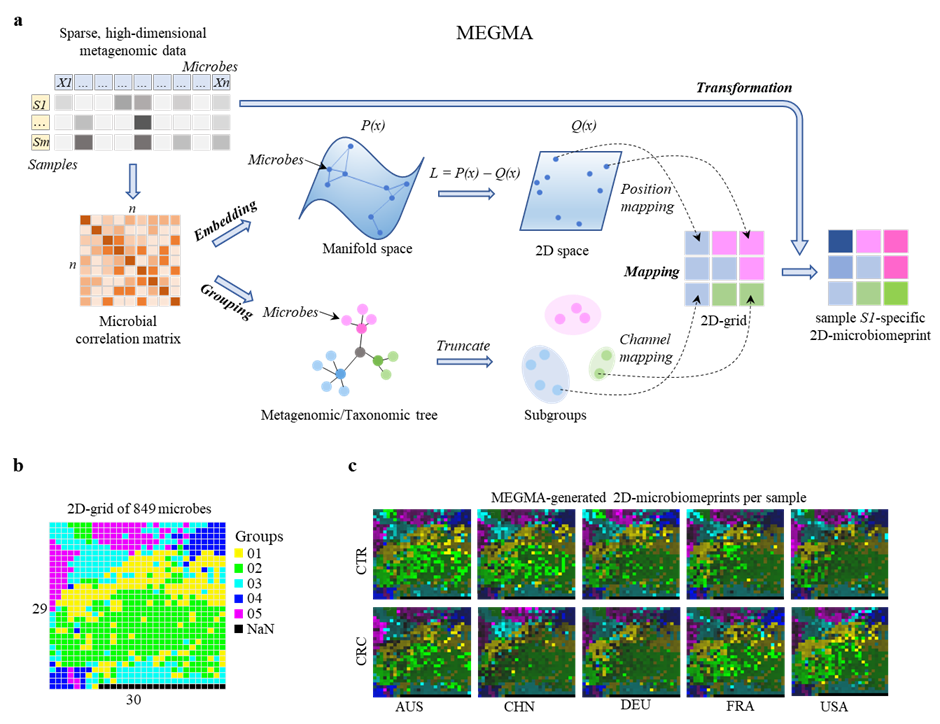
MEGMA is released in the aggmap package, in this example, we will show how to employ the aggmap package to perform MEGMA on the cross-nation datasets of metagenomics.
1.2 Metagenomic cross nation datasets and tasks
This metagenomic cross-nation data covers 849 gut microbial species from fecal samples of total 575 subjects (Colorectal cancers, CRCs or healthy controls, CTRs) compiled by Wirbel et al.(Nat. Med. 25, 679-689, 2019). It contains five metagenomic datasets from five separate studies in five nations (FRA: France, AUS: Australia, DEU: Germany, CHN: China, and USA: America).
Table 1. Metagenomic cross-nation data.
Dataset |
# Case (CRC) |
# Control (CTR) |
# Species |
Download |
|---|---|---|---|---|
AUS |
46 |
63 |
849 |
|
CHN |
74 |
54 |
849 |
|
DEU |
60 |
60 |
849 |
|
FRA |
53 |
61 |
849 |
|
USA |
52 |
52 |
849 |
The tasks are to build classfication models to classify CRCS from CTRs, and to identify the important microbial species that contribute to CRCs. Each microbe is a feature point in the classfication model, therefore, we need to calculate the feature importance to get the importance score of the microbes. Since we have five independent datasets that from five countries, we can build the model on one country data and test the model performance on the rest of the countries, we called this a
study-to-study transfer (STST, model trained on one nation data and tested on the rest nations) experiment. The STST can tell us the real performance of the metagenomc machine learning models in the application of the external data. Therefore, in this example, the study-to-study transfer will be used for the evaluation of the performance of the metagenomic prediction models and the consistency of the across-nation biomarker discovery. Note that since MEGMA is an unsupervised learning
method, we can use all unlabeled metagenomic data to fit megma (AKA, to train megma). Using all unlabeled data makes it more accurate and robust when calculating pairwise distances between microbes. Because the feature points in a country’s metagenomic data may have very low variance, the correlation between microbes cannot be accurately calculated.
1.3 MEGMA fitting and AggMapNet training strategy
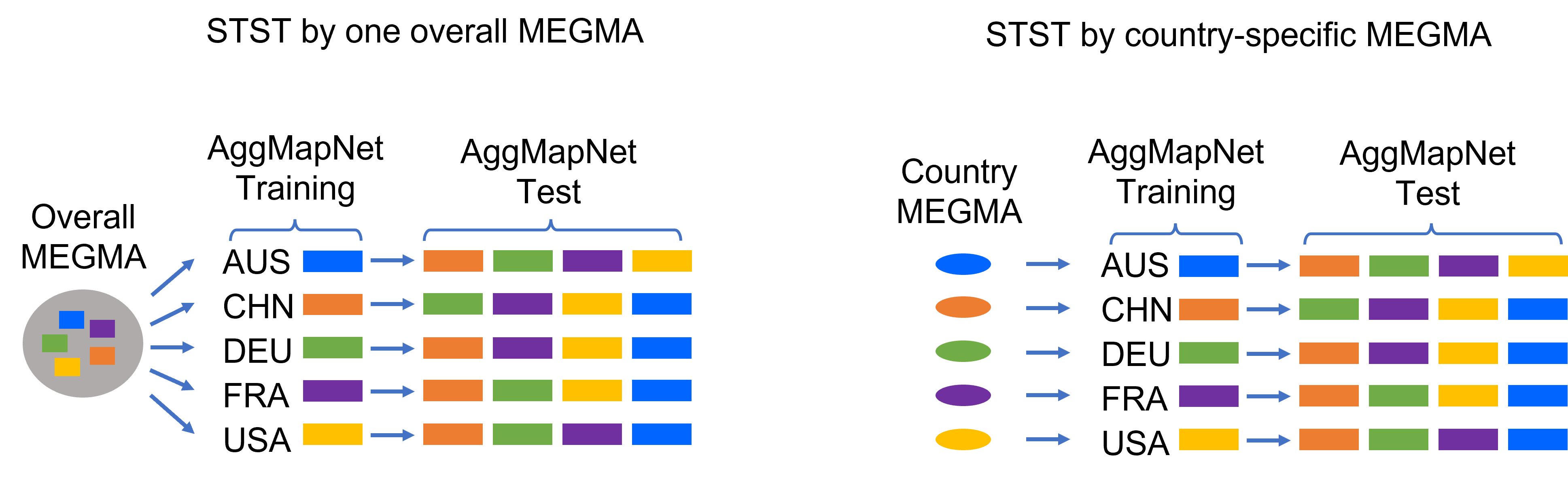
Because MEGMA is unsupervised learning method, therefore, in this stduy, we can use all of the metagenomic unlabelled abudance data to pre-fit MEGMA, we named this MEGMA as megma_overall. As shown in the figure below, the megma_overall can be fitted on the abundance data of the 5 countries: AUS, CHN, FRA, DEU, and USA. After that, the megma_overall can be used to transform the Fmaps for each country, and then we can build the AggMapNet based on the generated Fmaps of each country.
Except for that, we can also try to use unlabelled abudance data of one country only to fit the megma, for example if we use the USA abundance data, we can named our megma as megma_usa. We called this as country-specific megma, because megma is fitted on each country only. In the STST test stage, the Fmaps for the rest countries will be transformed from this megma. The country-specific megma maybe not as robust as the overall megma, because the samples to fit the country-specific
megma is lower than the samples to fit the overall megma.